library(ESRM64503)
library(kableExtra)
library(tidyverse)
library(DescTools) # Desc() allows you to quick screen data
library(lavaan) # Desc() allows you to quick screen data
# options(digits = 3)
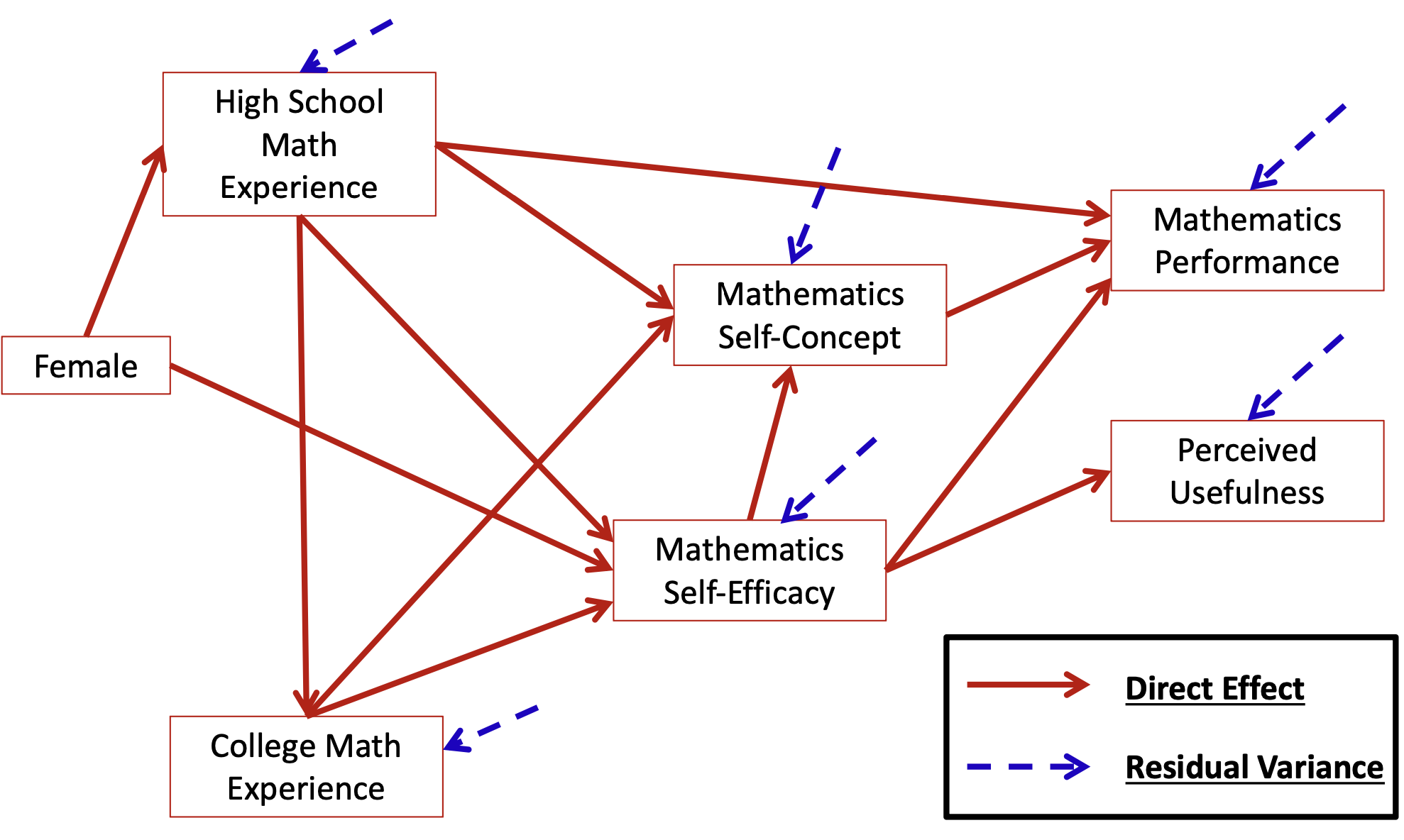
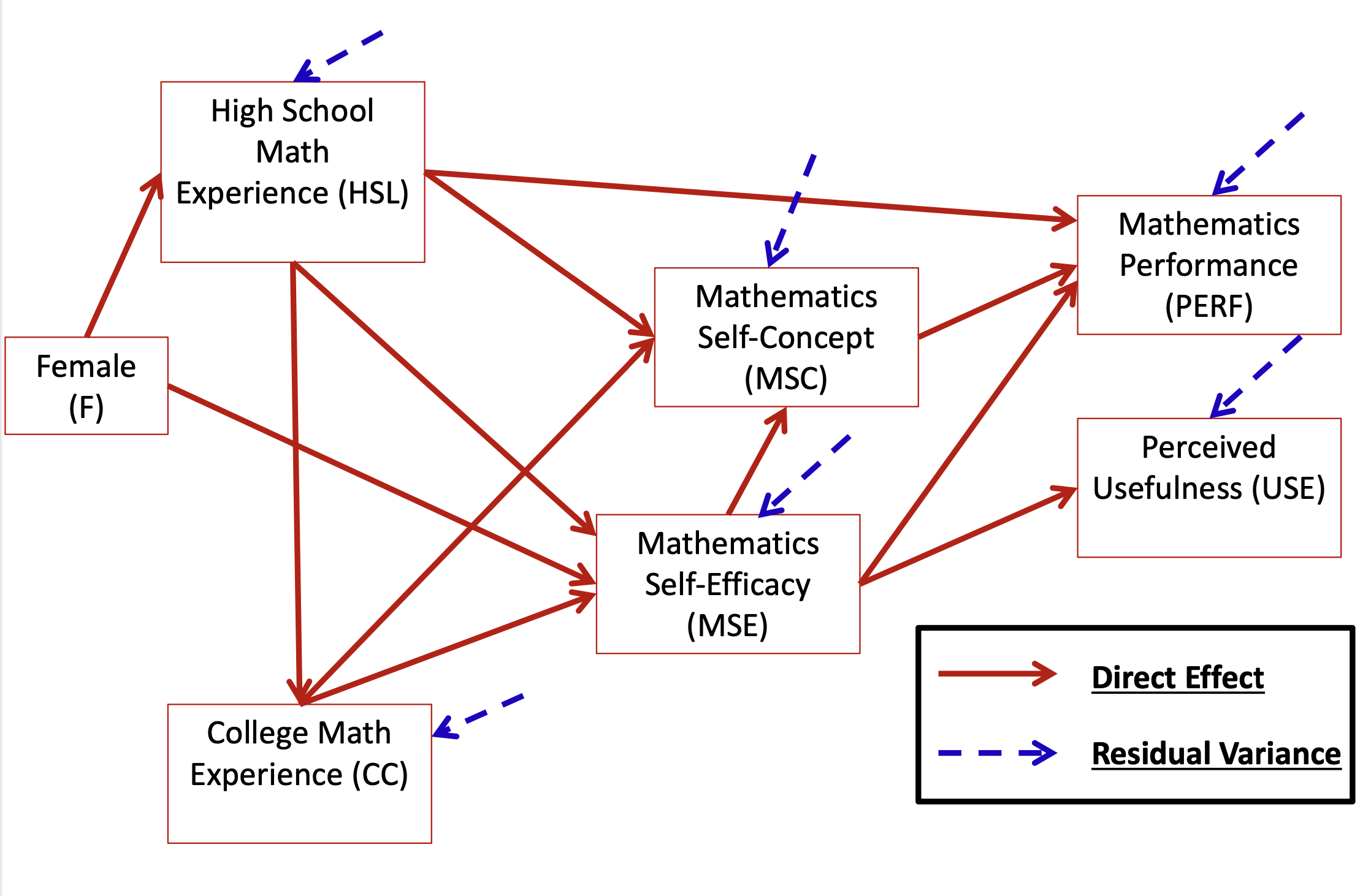
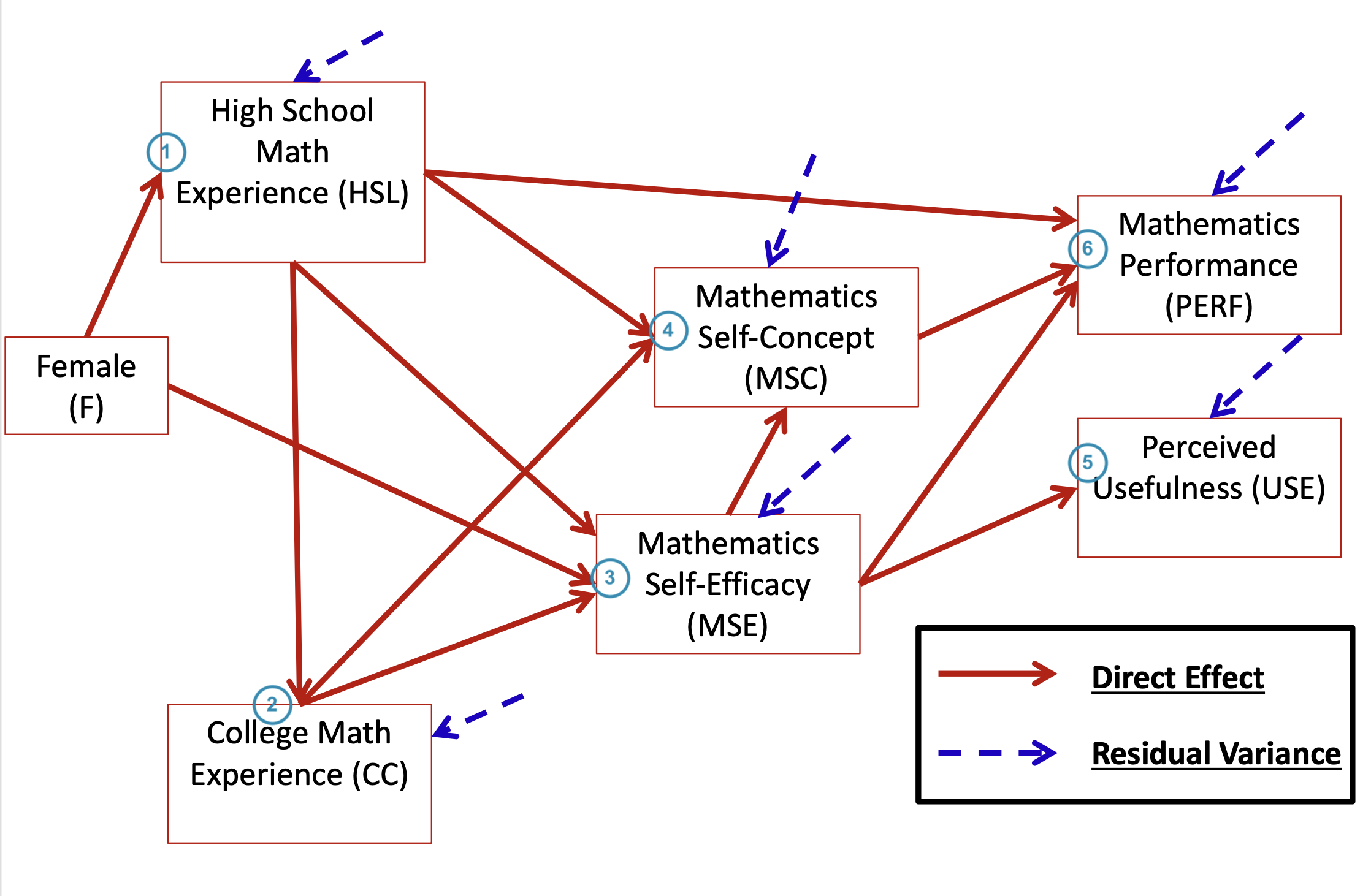
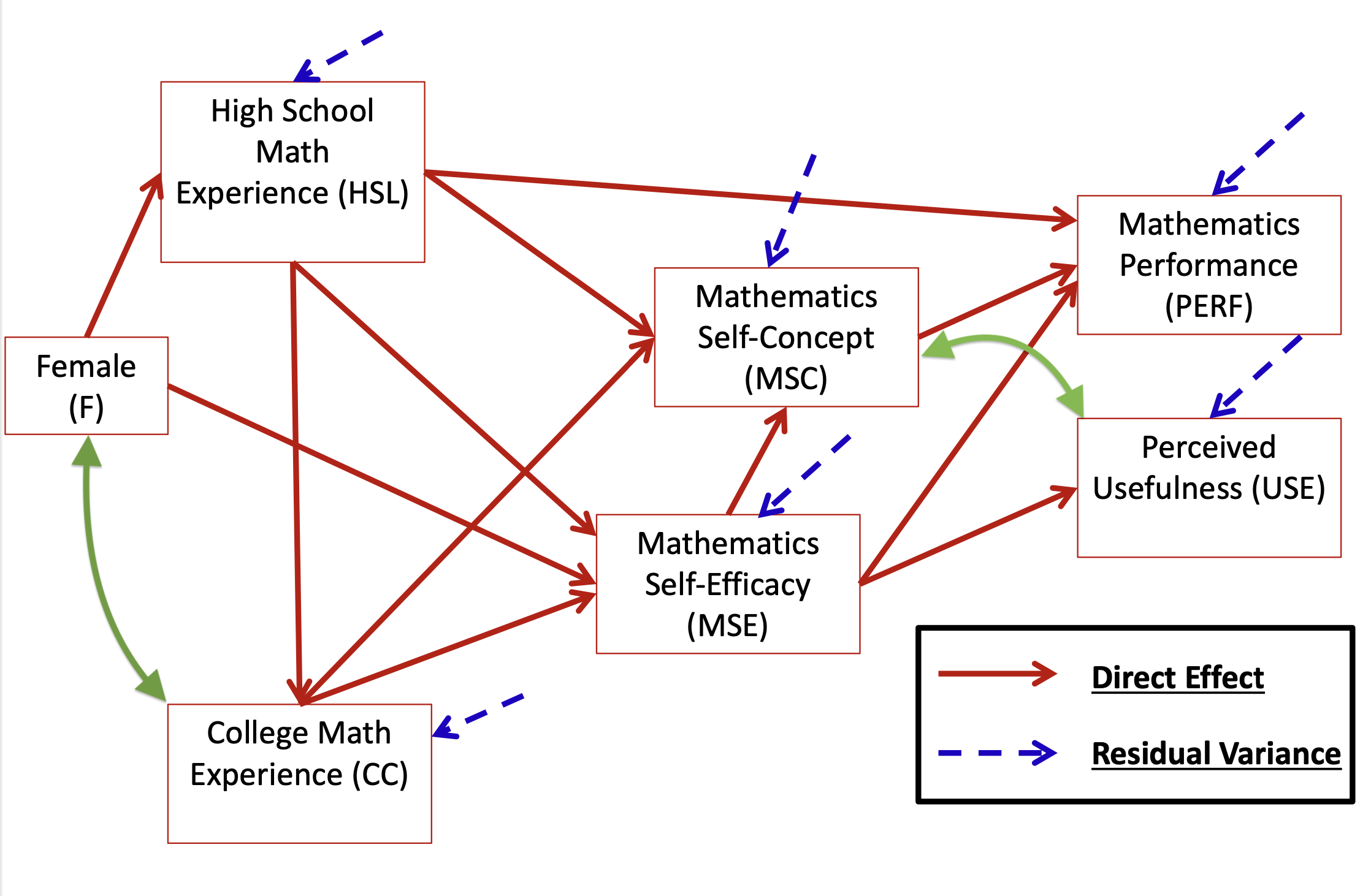
head(dataMath) id hsl cc use msc mas mse perf female
1 1 NA 9 44 55 39 NA 14 1
2 2 3 2 77 70 42 71 12 0
3 3 NA 12 64 52 31 NA NA 1
4 4 6 20 71 65 39 84 19 0
5 5 2 15 48 19 2 60 12 0
6 6 NA 15 61 62 42 87 18 0[1] 350 9